TO THE EDITOR—The outbreak of the coronavirus disease 2019 (COVID-19) pandemic due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was declared a pandemic on 12 March 2020 by the World Health Organization [1]. A major issue related to the outbreak has been to correlate viral RNA load obtained after reverse-transcription polymerase chain reaction (RT-PCR) and expressed as the cycle threshold (Ct) with contagiousness and therefore duration of eviction from contacts and discharge from specialized infectious disease wards. Several recent publications, based on more than 100 studies, have attempted to propose a cutoff Ct value and duration of eviction, with a consensus at approximately Ct >30 and at least 10 days, respectively [2–5]. However, in an article published in Clinical Infectious Diseases, Bullard et al reported that patients could not be contagious with Ct >25 as the virus is not detected in culture above this value [6]. This limit was then evoked in the French media during an interview with a member of the French Scientific Council Covid-19 as a possible value above which patients are no longer contagious [7].
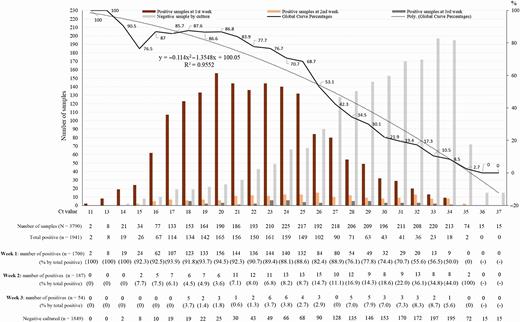
At the beginning of the outbreak, we correlated Ct values obtained using our PCR technique based on amplification of the E gene and the results of the culture [8]. Since the beginning of the pandemic, we have performed 250 566 SARS-CoV-2 RT-PCR for 179 151 patients, of whom 13 161 (7.3%) tested positive. Up to the end of May, 3790 of these samples, reported as positive on nasopharyngeal samples, were inoculated and managed for culture as previously described [8]. Of these 3790 inoculated samples, 1941 SARS-CoV-2 isolates could be obtained after the first inoculation or up to 2 blind subcultures. The correlation between the scanner values and the positivity of the culture allows us to observe that the image obtained with 10 times more isolates than in our preliminary work (1941 vs 129) does not change significantly (Figure 1). It can be observed that at Ct = 25, up to 70% of patients remain positive in culture and that at Ct = 30 this value drops to 20%. At Ct = 35, the value we used to report a positive result for PCR, <3% of cultures are positive. Our Ct value of 35, initially based on the results obtained by RT-PCR on control negative samples in our laboratory and initial results of cultures [8], is validated by the results herein presented and is in correlation with what was proposed in Korea [9] and Taiwan [10]. We could observe that subcultures, especially the first one, allow an increasing percentage of viral isolation in samples with Ct values, confirming that these high Ct values are mostly correlated with low viral loads. From our cohort, we now need to try to understand and define the duration and frequency of live virus shedding in patients on a case-by-case basis in the rare cases when the PCR is positive beyond 10 days, often at a Ct >30. In any cases, these rare cases should not impact public health decisions.
Notes
Ethical approval. The protocol was approved by the University Hospital Institute Méditerranée Infection Ethical Committee. All patients provided informed consent in accordance with the Declaration of Helsinki.
Financial support. This research was funded by the French Government under the Investissements d’avenir (Investments for the Future) program managed by the Agence Nationale de la Recherche (French National Agency for Research; reference: Méditerranée Infection 10-IAHU-03) and by Région Provence-Alpes-Côte d’Azur and European funding Fond Europen de Recheche et de developpement (FEDER) PRIMI.
Potential conflicts of interest. D. R. reports grants from the Hitachi High-Tech Corporation outside the submitted work. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
1.Adhanom 2020.World Health Organization director-general’s opening remarks at the media briefing on COVID-19—11 March 2020. Available at: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19—11-march-2020.
Accessed 20 September 2020.
2.Jefferson T,Spencer E,Brassey J, Heneghan C.Viral cultures for COVID-19 infectivity assessment. Systematic review. medRxiv 2020: 2020.08.04.20167932.Google Scholar
3.Rhee C,Kanjilal S,Baker M,Klompas M.Duration of SARS-CoV-2 infectivity: when is it safe to discontinue isolation?Clin Infect Dis 2020; doi:10.1093/cid/ciaa1249.Google Scholar
4.Singanayagam A, Patel M,Charlett A,et al.Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020.
Eurosurveillance 2020; 25:2001483. doi: 10.2807/1560-7917.ES.2020.25.32.2001483.
5.Cevik M, Tate M,Lloyd O, Maraolo AE, Schafers J, Ho A. SARS-CoV-2, SARS-CoV-1 and MERS-CoV viral load dynamics, duration of viral shedding and infectiousness: a living systematic review and meta-analysis. Infectious Diseases (except HIV/AIDS) 202. doi: 10.1101/2020.07.25.20162107.Google Scholar
6.Bullard J, Dust K, Funk D, et al.Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis 2020; doi:10.1093/cid/ciaa638. Google Scholar
7.Senecat A . Covid-19 : l’hypersensibilité des tests PCR, entre intox et vrai débat. Le Monde.fr. 2020. Available at: https://www.lemonde.fr/les-decodeurs/article/2020/09/09/covid-19-l-hypersensibilite-des-tests-pcr-entre-intox-et-vrai-debat_6051528_4355770.html. Accessed 20 September 2020
8.La Scola B, Le Bideau M, Andreani J, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis 2020; 39:1059–61.Google Scholar Crossref
9.Chang MC, Hur J, Park D. Interpreting the COVID-19 test results: a guide for physiatrists . Am J Phys Med Rehabil 2020. doi:10.1097/PHM.0000000000001471.
10.Chen CJ, Hsieh LL, Lin SK, et alOptimization of the CDC protocol of molecular diagnosis of COVID-19 for timely diagnosis. Diagnostics 2020; 10:333.Google Scholar
Crossref © The Author(s) 2020. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail: journals.permissions@oup.com.
This article is published and distributed under the terms of the Oxford University Press, Standard Journals Publication Model (https://academic.oup.com/journals/pages/open_access/funder_policies/chorus/standard_publication_model)
https://academic.oup.com/